Areas of Research: Microbial pathogenesis, protozoan parasites, viruses of protozoan parasites, glycobiology, genetic crossing and genomics employing CRISPR gene editing, biochemistry and cell biology, host responses and immunology
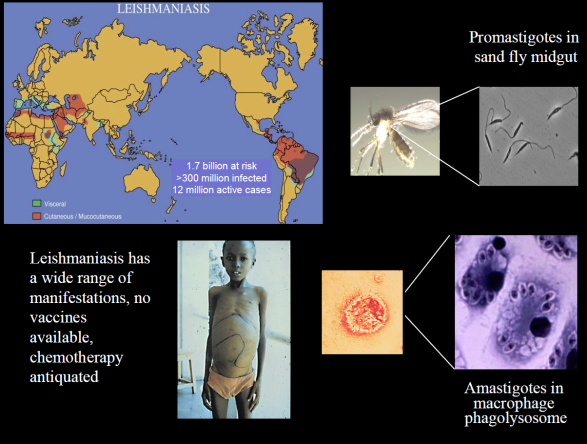
Much of our work focuses on the protozoan parasite Leishmania, infecting more than 10 million people in tropical regions. We want to understand how the parasite carries out its infectious cycle, which comprises an intracellular phagolysosomal stage within vertebrate macrophages and an extracellular stage within the gut of a sand fly vector.
The development and application of new methods to parasite virulence.
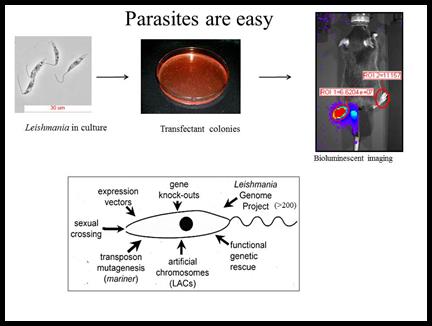
Experimentally, Leishmania is EASY to work with. Genetic manipulations are fast and routine, and one can plate them and generate many independent colonies. Parasites can be grown readily in suspension culture under conditions where they differentiate well, and there are excellent mouse models enabling dissection of host defenses.
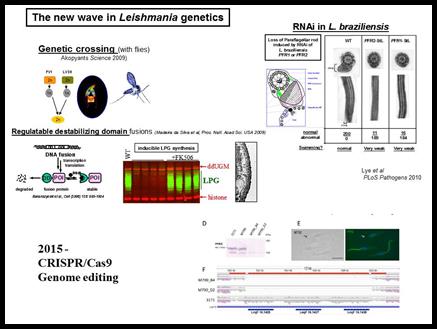
We have developed and/or applied in parasites powerful methods such as functional genetic rescue, gene knockouts, transposon mutagenesis, expression profiling via microarrays or RNA sequencing, and genome sequencing. We have identified many genes with ‘interesting’ functions, expression patterns and/or database relationships, and now are in the process of assessing their role in virulence using a variety of genetic and biochemical methods. One of the challenges of Leishmania is diploidy and an as yet inconvenient sexual cycle in the sand fly vector. The advent of RNAi tools and more recently CRISPR technology readily overcomes this bottleneck. CRISPR has many advantages and we have achieved very high efficiencies of CRISPR editing without need for selection. Notably these can be applied not only to the parasite but also to the host genomics, to probe host-parasite interactions from both perspectives.
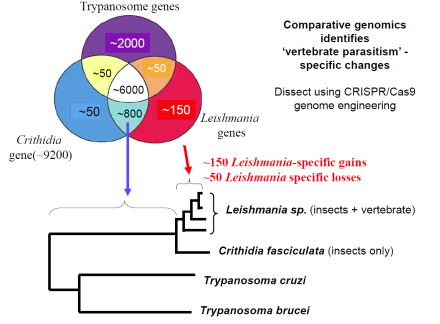
Functional genomics.
In 2009 we showed for the first time that genetic crossing is possible in Leishmania. This opens the way to genetic analysis of important traits including tropisms related to different forms of leishmaniasis (cutaneous, mucocutaneous or visceral disease). While crossing has thus far been seen only within the sand fly vector, some data suggest that in vitro crossing may be possible which would greatly facilitate these studies. We are resequencing several Leishmania genomes to provide SNP markers for this purpose, as well as to create the first Leishmania genetic map. In conjunction with an NHGRI/NIAID sponsored white paper initiative we are working with a consortium of scientists and the Genome Sequencing Center of Washington University to sequence over 50 new trypanosomatid genomes. Comparative analysis will permit identification of genes associated with phylogenetically with different manifestations of Leishmania pathology and virulence for functional testing.
Parasite Virology!
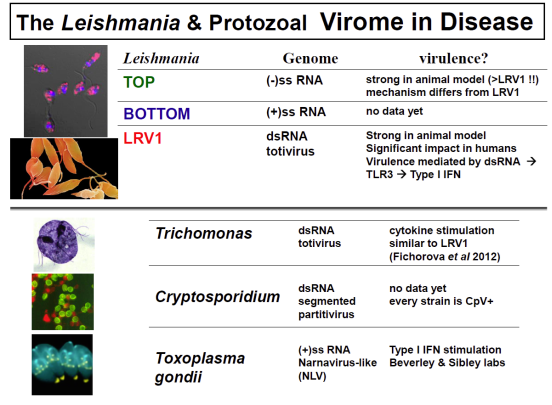
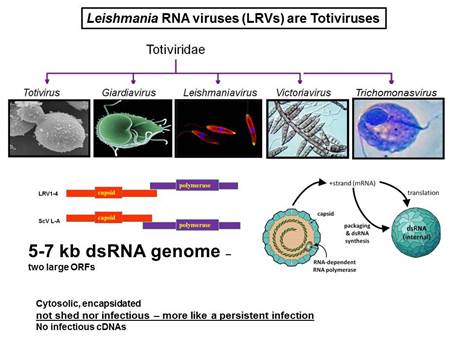
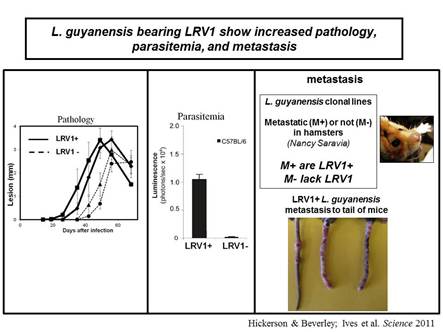
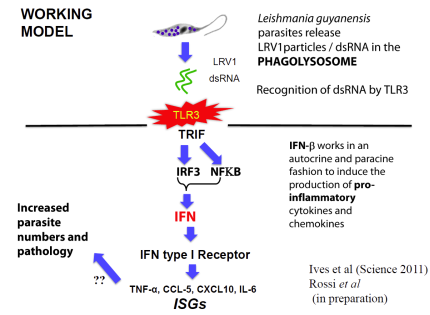
In collaboration with the group of Nicolas Fasel in Lausanne Switzerland, we showed in 2011 that enigmatic dsRNA Leishmaniavirus (LRV) was associated causally with increased virulence and increased metastasis. LRV-infected Leishmania induce a hyperimmune response to the parasite responsible for the increased pathogenicity, through a dsRNA and TLR3-dependent pathway. We are focusing now on the mechanism by which this occurs, which includes Type I interferon signaling as a key player.
Notably, L. braziliensis and related species of Viannia are responsible for a severe form of leishmaniasis called mucocutaneous leishmaniasis; the LRV represents the first parasite-associated factor implicated in this terrible disease. Our groups are now working to together to better understand how and where LRV contributes to human disease. We are surveying strains across South America for LRVs in an effort to associate LRV with human disease manifestations, and developing experimental Leishmania LRV ‘virology’ to assess in the laboratory how LRV contributes to pathology. The role of RNA interference in LRV biology and LRV-dependent virulence is a subject of considerable interest as well.
A new focus is how general the ‘hypervirulence’ associated with parasite viruses may be; a great many have been described in other parasites but like LRV1, previously overlooked. We are pursuing known viral elements as carrying out viral discovery efforts, not only in Leishmania (with the discovery of several new viruses) but also in Trichomonas vaginalis (with Pat Johnson at UCLA), Cryptosporidium (with Mark Kuhlenschmidt at the University of Illinois), and Toxoplasma (with David Sibley in the Molecular Microbiology department). Current data are encouraging.
The Leishmania surface coat and its interface with the host.
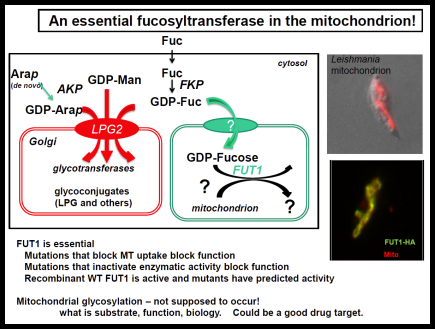
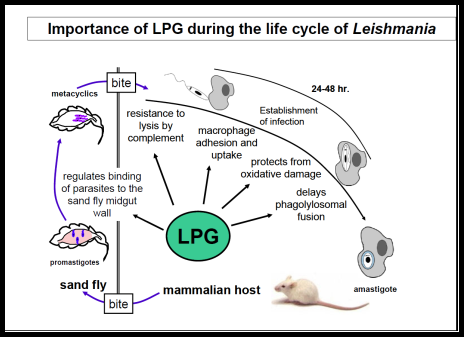
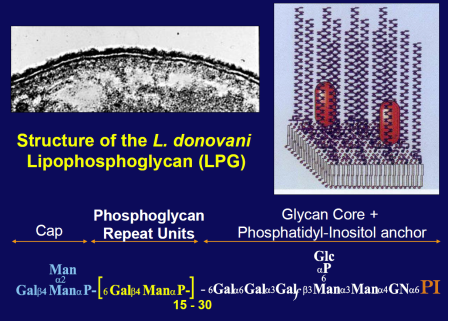
The Leishmania surface is composed of a dense glycocalyx of glycolipids. One fruitful genetic screen involves a surface glycoconjugate, lipophosphoglycan (LPG), an essential virulence determinant involved in adhesion and survival. LPG genes encode proteins involved in biosynthesis, compartmentalization of LPG intermediates within the eukaryotic secretory pathway, and regulation. We are especially interested in understanding the role of the parasite surface interactions in manipulating host cell signal transduction, which is radically altered in Leishmania infections, and in sand fly transmission and co-evolution.
A role for the parasite flagellum in virulence.
While the infective amastigote stage lacks an external flagellum, it maintains a ‘stub’ which has largely been ignored. Recently Gull’s lab proposed this may play a role in parasite virulence, as a conduit for virulence factors as well as a sensing organelle as seen in other eukaryotes. To test this we sought to knockout the Leishmania flagellum. Remarkably this proved easy to do through ablation of a gene implicated in intraflagellar transport, IFT140. In culture as the promastigote stage, these parasites completely lacked a flagellum, yet maintained a fairly normal flagellar pocket, and grew normally (other than being unable to swim). In contrast, the ift140 mutants were completely avirulent, in a mouse or in a macrophage model. Current work is now directed towards understanding the impact of this on virulence factor secretion into the host.
RNA interference.
In 2010 we showed that while most Leishmania species lack the RNA interference pathway, species belonging to the L. braziliensis group have retained it. This permits the application of RNAi-based methods for functional genomics in this group of species, although the advent of CRISPR gene editing will likely supplant this going forward. The loss of RNAi in most Leishmania raises the interesting evolutionary question of what forces contribute to the loss of a pathway of such fundamental importance in other organisms. This is being pursued, through the study of RNAi-deficient null mutants many of which bear RNA viruses.
Other interests of the lab include molecular evolution of parasitism and virulence, and the use of genetically modified parasites as vaccines.